Run CONSTAX locally¶
This is a simple tutorial about CONSTAX. We will explain how to run CONSTAX on a local computer like a laptop or a desktop computer.
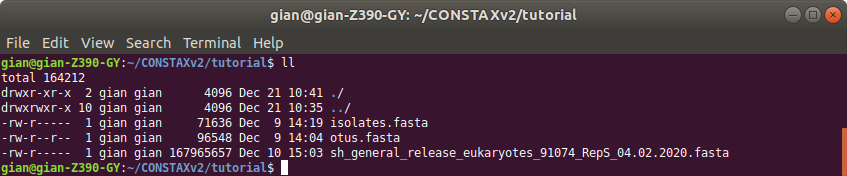
Before we start, we need to create a folder called tutorial. This CONSTAX test will happen
inside this folder so you first need to copy all the files you we will use before running the
software. We need the OTU representative sequence fasta file (e.g. otus.fasta),
the representative sequence fasta file of your culture isolates if you have any and you want to
try to match with the OTUs (e.g. isolates.fasta), and the sequence reference database you want
- to use, for Fungi (e.g.
sh_general_release_eukaryotes_91074_RepS_04.02.2020.fasta, see theSuggested Reference Databases page for details). These files must end in the extensions
.fasta,.fa, or.fna.
You tutorial folder should look like this:
It is smart to use the sh command line interpreter, so we will create a .sh file and write the CONSTAX commands in it.
gian@gian-Z390-GY:~/tutorial$ nano constax.sh
This is how the content of the .sh file should look like
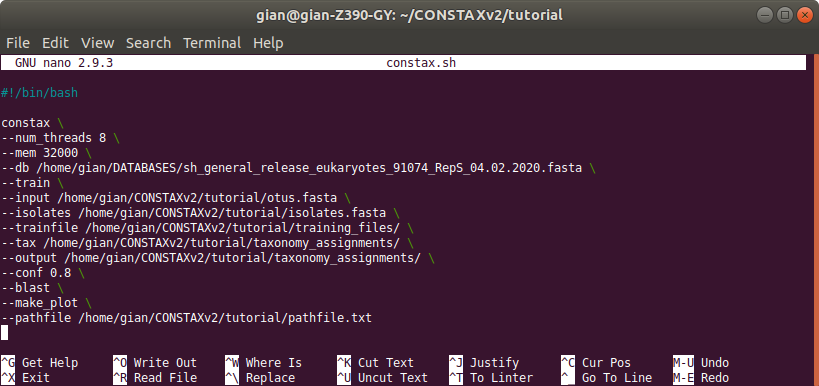
constax \
--num_threads 10 \
--mem 32000 \
--db /home/gian/DATABASES/sh_general_release_eukaryotes_91074_RepS_04.02.2020.fasta \
--train \
--input /home/gian/CONSTAXv2/tutorial/otus.fasta \
--isolates /home/gian/CONSTAXv2/tutorial/isolates.fasta \
--trainfile /home/gian/CONSTAXv2/tutorial/training_files/ \
--tax /home/gian/CONSTAXv2/tutorial/taxonomy_assignements/ \
--output /home/gian/CONSTAXv2/tutorial/taxonomy_assignements/ \
--conf 0.8 \
--blast \
--make_plot \
--pathfile /home/gian/CONSTAXv2/tutorial/pathfile.txt
Note
Remember. If using a reference database for the first time, you will need to use the -t or -\-train flag to train the classifiers on the dataset. The training step is necessary only at first use, you can just point to the -\-trainfile <PATH> for the subsequent classifications with the same reference database. For SILVA please see the Download and generate SILVA reference database page for details on how to create a valid SILVA database before running CONSTAX.
The --pathfile option is necessary ONLY if you are planning to use USEARCH instead of VSEARCH for your classification. In this case we suggested to create a pathfile.txt
gian@gian-Z390-GY:~/tutorial$ nano pathfile.txt
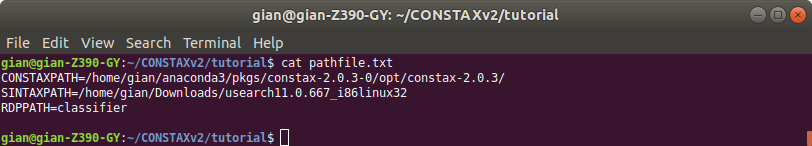
where you will add the absolute PATHs for the required software. VSEARCH, BLAST, and RDP are already available through the conda environment, what you will need is just USEARCH for the SINTAX classification. The pathfile.txt should look like this below:
Warning
Remember to navigate through your anaconda installation and find the constax-2.0.17/ folder.
This is the only way to make CONSTAX locate the needed python scripts.
Before you can run CONSTAX you need to activate your anaconda environment (alternatively, you can include this in the constax.sh file).
gian@gian-Z390-GY:~/tutorial$ conda activate
To see how to set up a conda environment with CONSTAX please refer to this link.
At this point your are ready to give CONSTAX a try.
gian@gian-Z390-GY:~/tutorial$ constax
And CONSTAX will start running…
When CONSTAX will be done you will see the outputs in the working directory.
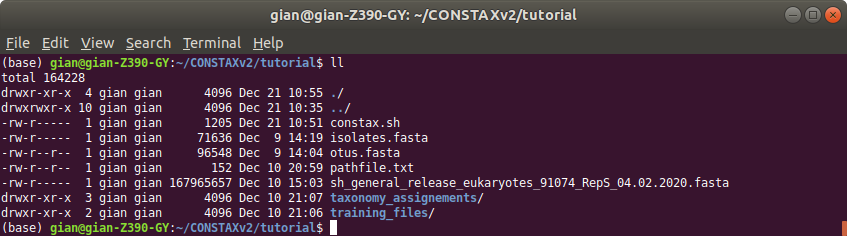
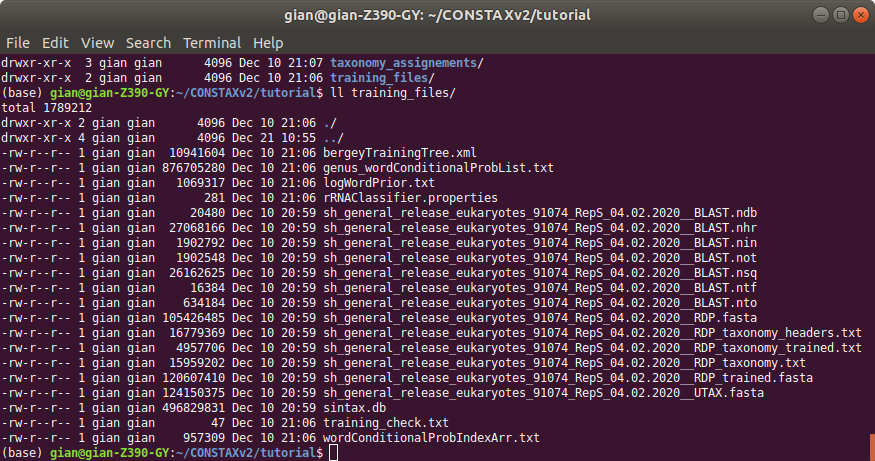
Training file and classification results will be stored in the specified folders. In this example
the training files will be in training_files
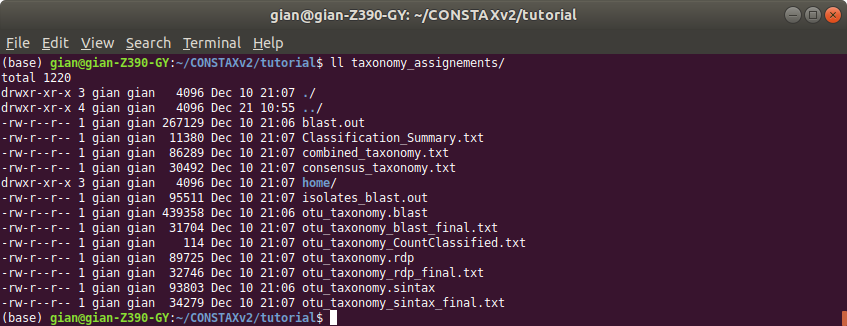
and the classification in taxonomy_assignments
The taxonomic classification of your OTUs representative sequences will be in constax_taxonomy.txt.
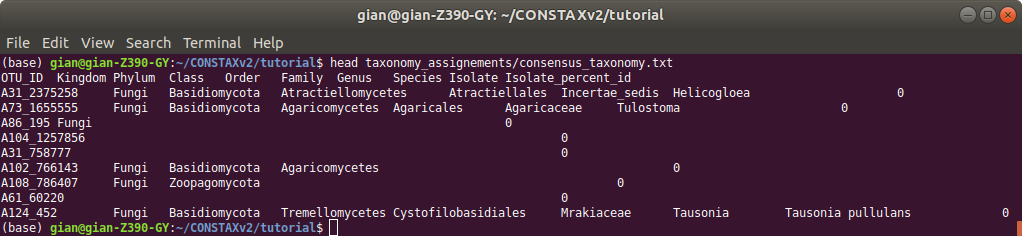
While classifications performed by each classifier will be store in combined_taxonomy.txt
Please explore other CONSTAX outputs, such as Classification_Summary.txt.
If you want to use some test otus.fasta to practice the use of CONSTAX you can find some in THIS github repo of CONSTAX.
Now. We can try to run CONSTAX again changing some parameters to see some other options.
For example, modify the constax.sh script as showed below.