Download and generate SILVA reference database¶
This is a tutorial about how to generate a reference database, that can be used with CONSTAX. from the SILVA database that contains Bacteria and Archaea sequences.
First thing to do is to download the SILVA reference database here.
You should use the latest release such as 138. Go to release_<XXX> > Exports where <XXX> is the release number, and download a gzipped fasta such as SILVA_138_SSURef_tax_silva.fasta.gz
with the name ending in _SSURef_tax_silva.fasta.gz.
wget https://www.arb-silva.de/fileadmin/silva_databases/release_138/Exports/SILVA_138_SSURef_tax_silva.fasta.gz
gunzip SILVA_138_SSURef_tax_silva.fasta.gz
curl -O https://www.arb-silva.de/fileadmin/silva_databases/release_138/Exports/SILVA_138_SSURef_tax_silva.fasta.gz
gunzip SILVA_138_SSURef_tax_silva.fasta.gz
Then, the best way is to create a script (it can be and .sh file or a .sb file depending
if you are running CONSTAX locally or on the MSU HPCC) that generates the Bacteria and the Archaea
fasta files and directly concatenate them together.
This is how the content of the .sh file should look like
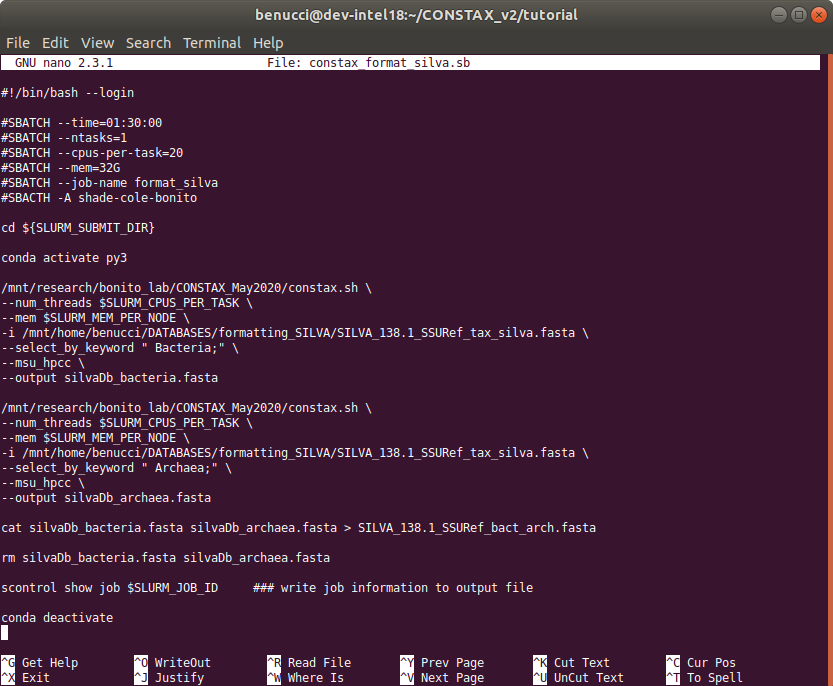
You can copy and paste this code below as a guideline.
#!/bin/bash
constax \
-i SILVA_138_SSURef_tax_silva.fasta \
--select_by_keyword " Bacteria;" \
--output silva_Db_bacteria.fasta
constax \
-i SILVA_138_SSURef_tax_silva.fasta \
--select_by_keyword " Archaea;" \
--output silva_Db_archaea.fasta
cat silva_Db_bacteria.fasta silva_Db_archaea.fasta > SILVA_138_SSURef_bact_arch.fasta
rm silva_Db_bacteria.fasta silva_Db_archaea.fasta
Warning
Remember to specify the keywords correctly, as they appear in the SILVA reference.
For example, to target the domain Bacteria the right keyword is " Bacteria;"
with a space before the name and “;” after it.
When the scripts are finished running you can inspect the results.
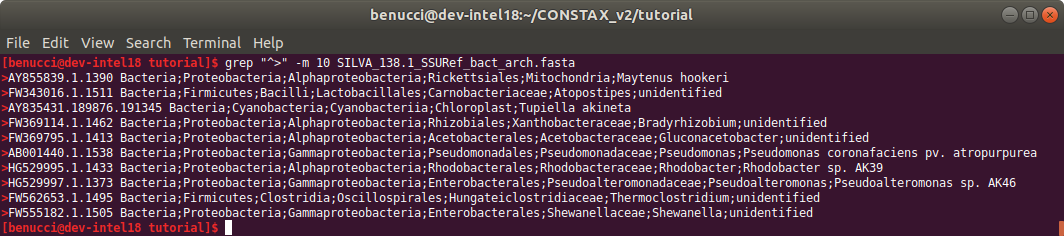
grep "^>" -m 10 SILVA_138.1_SSURef_bact_arch.fasta
The headers are formatted correctly and you can now use the newly created reference to classify your sequences.