Run CONSTAX on HPCC¶
To run CONSTAX on the high performance cluster computer or HPCC
available at Michigan State University, you can set the paths just using --msu_hpcc
flag to your constax.sh file
The code will look like as below
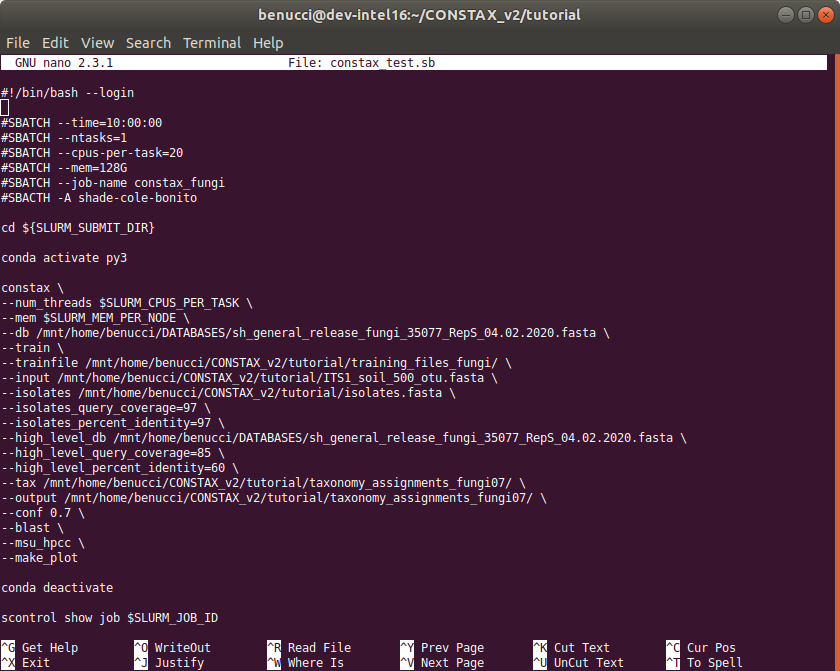
#!/bin/bash --login
#SBATCH --time=10:00:00
#SBATCH --ntasks=1
#SBATCH --cpus-per-task=20
#SBATCH --mem=32G
#SBATCH --job-name constax_fungi
#SBACTH -A shade-cole-bonito
cd ${SLURM_SUBMIT_DIR}
conda activate py3
constax \
--num_threads $SLURM_CPUS_PER_TASK \
--mem $SLURM_MEM_PER_NODE \
--db /mnt/home/benucci/DATABASES/sh_general_release_fungi_35077_RepS_04.02.2020.fasta \
--train \
--trainfile /mnt/home/benucci/CONSTAX_v2/tutorial/training_files_fungi/ \
--input /mnt/home/benucci/CONSTAX_v2/tutorial/ITS1_soil_500_otu.fasta \
--isolates /mnt/home/benucci/CONSTAX_v2/tutorial/isolates.fasta \
--isolates_query_coverage=97 \
--isolates_percent_identity=97 \
--high_level_db /mnt/home/benucci/DATABASES/sh_general_release_fungi_35077_RepS_04.02.2020.fasta \
--high_level_query_coverage=85 \
--high_level_percent_identity=60 \
--tax /mnt/home/benucci/CONSTAX_v2/tutorial/taxonomy_assignments_fungi07/ \
--output /mnt/home/benucci/CONSTAX_v2/tutorial/taxonomy_assignments_fungi07/ \
--conf 0.7 \
--blast \
--msu_hpcc \
--make_plot
conda deactivate
scontrol show job $SLURM_JOB_ID
Note
As you can see this time constax.sh does not contain the --train option,
since the reference database has been already trained it is not required any
additional training. This will improve the speed and therefore the running time
will be less. The resources you need to compute just the classification are much
less that those needed for training. You can then set the num_threads option
to a lower number as well as the amount of RAM --mem.
Additionally no --isolates is provided in this run of CONSTAX and the --hpcc_msu
is specified at the end of the script.
To access some other representative OTU sequences files please follow THIS link. These are the available files.